DNA Cardiovascular Health Test
We offer two types of tests; Lab Tests and Rapid Tests. This product is under the category Lab Tests. See all our Lab Tests by following the link.
See allWe offer several different options of testing methods. This test is done with Saliva. See all tests done with Saliva by following the link.
See allThe DNA Cardiovascular Health test provides clarity on how your genes affect the heart and blood vessels. This DNA test analyzes genes related to heart rate, recovery, and other factors important for cardiovascular health, offering you specific recommendations based on your results.
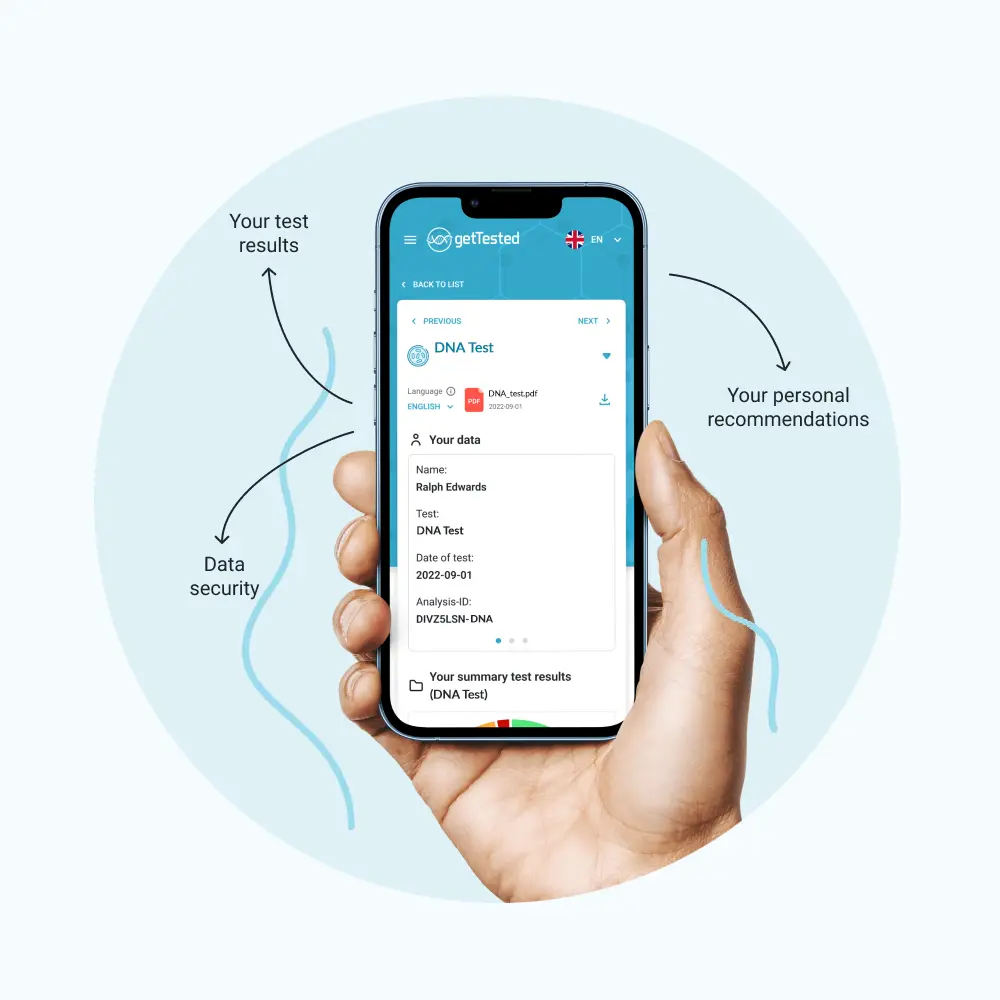
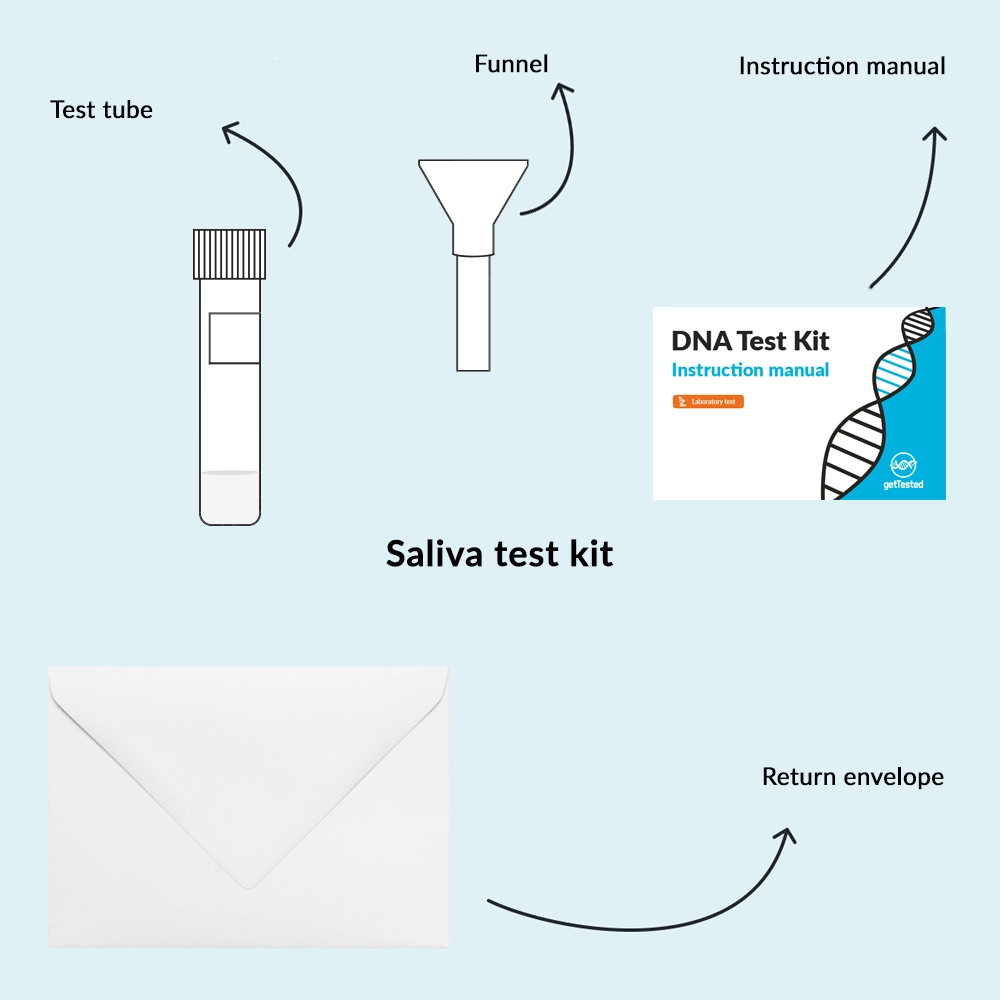
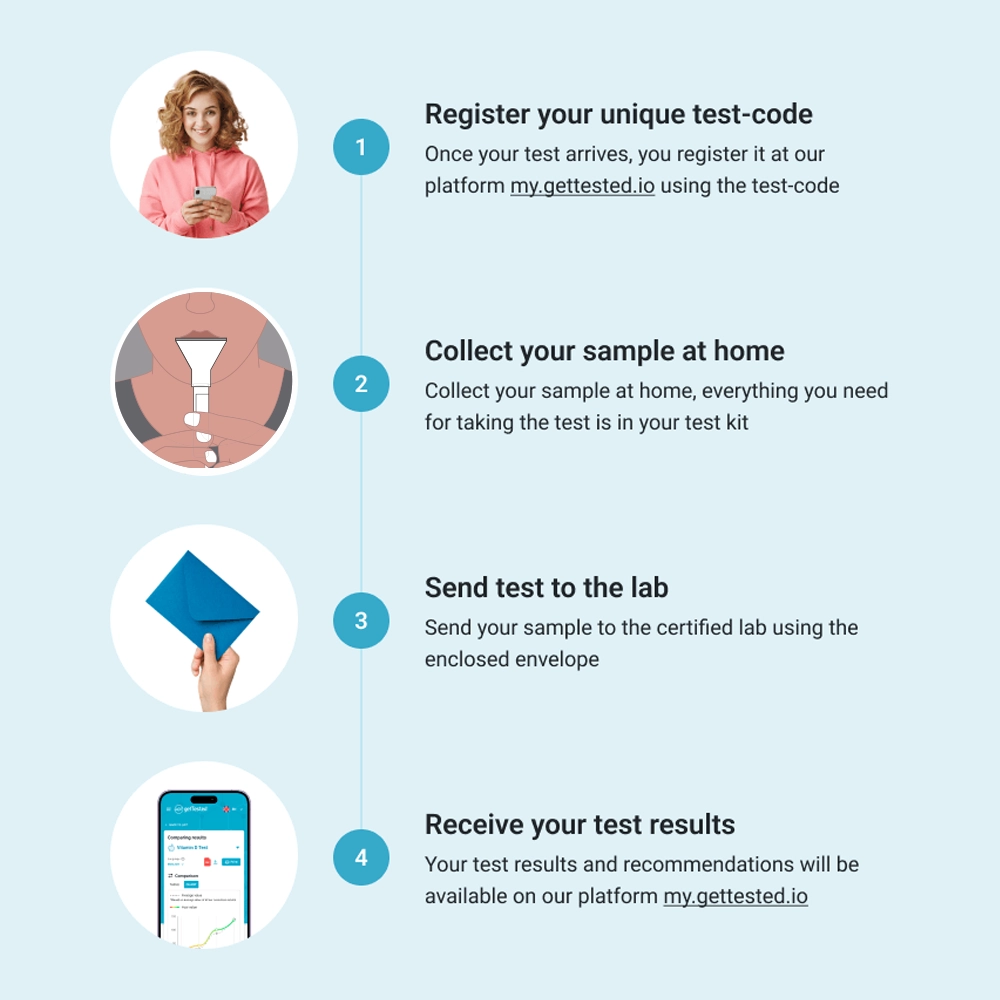
You can easily collect your saliva sample with the included test kit and send it to our lab. The price covers the return shipping. You’ll receive your detailed results digitally within 6-8 weeks.
Save more by bundling DNA tests. Order multiple tests together for better value from our wide range.
- In stock
- At-home test
- Results 6-8 weeks

Get 5% off on 2 Lab tests, and 10% off on 3 Lab tests or more.
What is analyzed in the test?
Heart Rate
Heart Rate Variability
Heart Rate Recovery
Salt Sensitivity
ApoB
TMAO
Homocysteine
LDL Particle Size
FAQ
How is the DNA Cardiovascular Health test carried out?
How quickly will I receive my results?
When should I take the test?
Example Report
Example of DNA Cardiovascular Health Test
Reviews
-
Could you please confirm if the cardiovascular test will cover hypertension and the consequences of and if not do you do such a test?Hi Rowena! Sorry to keep you waiting. These comments sometimes get lost in all the spam comments. If you email us at [email protected], I can send you a sample report for you to see. With that said, it doesn't directly test for your risk of hypertension. However, many of the genes affecting cardiovascular disease are indirectly involved in hypertension as well.
Related Products
-
Cholesterol Test
Test your cholesterol levels with GetTested’s Cholesterol Test Kit. Many people hav...€ 39,00 Add to cart -
D-dimer test (blood clots)
The GetTested D-dimer Rapid Test offers a rapid and accurate way to assess D-dimer le...€ 24,99 Add to cart -
DNA Combo 3 - Order any 3 tests
You can combine any of our DNA tests to get a discount. Combine any 3 reports for EUR...Original price was: € 499,00.€ 325,00Current price is: € 325,00. Add to cart -
Estrogen & progesterone test
GetTested's Estrogen & Progesterone Test measures the absolute levels and the rat...€ 79,00 Add to cart
You may also like…
Trusted by over 10.000+ customers


“The home test was straightforward with easy to follow instructions. The test result was detailed and clear in its presentation. Also had the opport...”
Richard

“We looked at a lot of companies offering the same services but made the decision to go forward with get tested because the labs are based here in t...”
Natasha

“There are other providers out there, I have tried 3. Gettested was the fastest and the customer service was the best. I even received my test on th...”
Alan

“Absolutely perfect, results came after few days and finally after a lots of times visited GP, we finally know why my son has eczema - he is allergy...”
E

“I found them to be professional and not too long a wait for results. Helpful with any after questions. Will be using them again if need be. I highl...”
Eileen

“I really value my test and to see the results. It is a jungle when your health is online. So get accurate and precise knowledge is a gift, as it is...”




























Leave a Reply